
What is the cloverleaf model of tRNA? Structure, Diagram, Function Explained
The concept of Short Notes Of Biology For NEET tRNA Structure is essential in biology and helps explain real-world biological processes, especially protein synthesis. Mastering tRNA structure is vital for solving NEET exam questions based on molecular genetics, translation, and cell biology.
Understanding Short Notes Of Biology For NEET tRNA Structure
Short Notes Of Biology For NEET tRNA Structure refers to concise, exam-oriented explanations covering the structure, diagram, and function of transfer RNA (tRNA). This topic is important in areas like translation process, protein synthesis, and genetic code decoding. Understanding the secondary (cloverleaf) and tertiary (L-shaped) structure of tRNA is key for NEET aspirants targeting cell biology and molecular inheritance MCQs.
Here’s a helpful table to understand Short Notes Of Biology For NEET tRNA Structure better:
Short Notes Of Biology For NEET tRNA Structure Table
| Level/Type | Description | Key Points |
|---|---|---|
| Primary Structure | Linear sequence of 76–90 ribonucleotides | Sequence determines function |
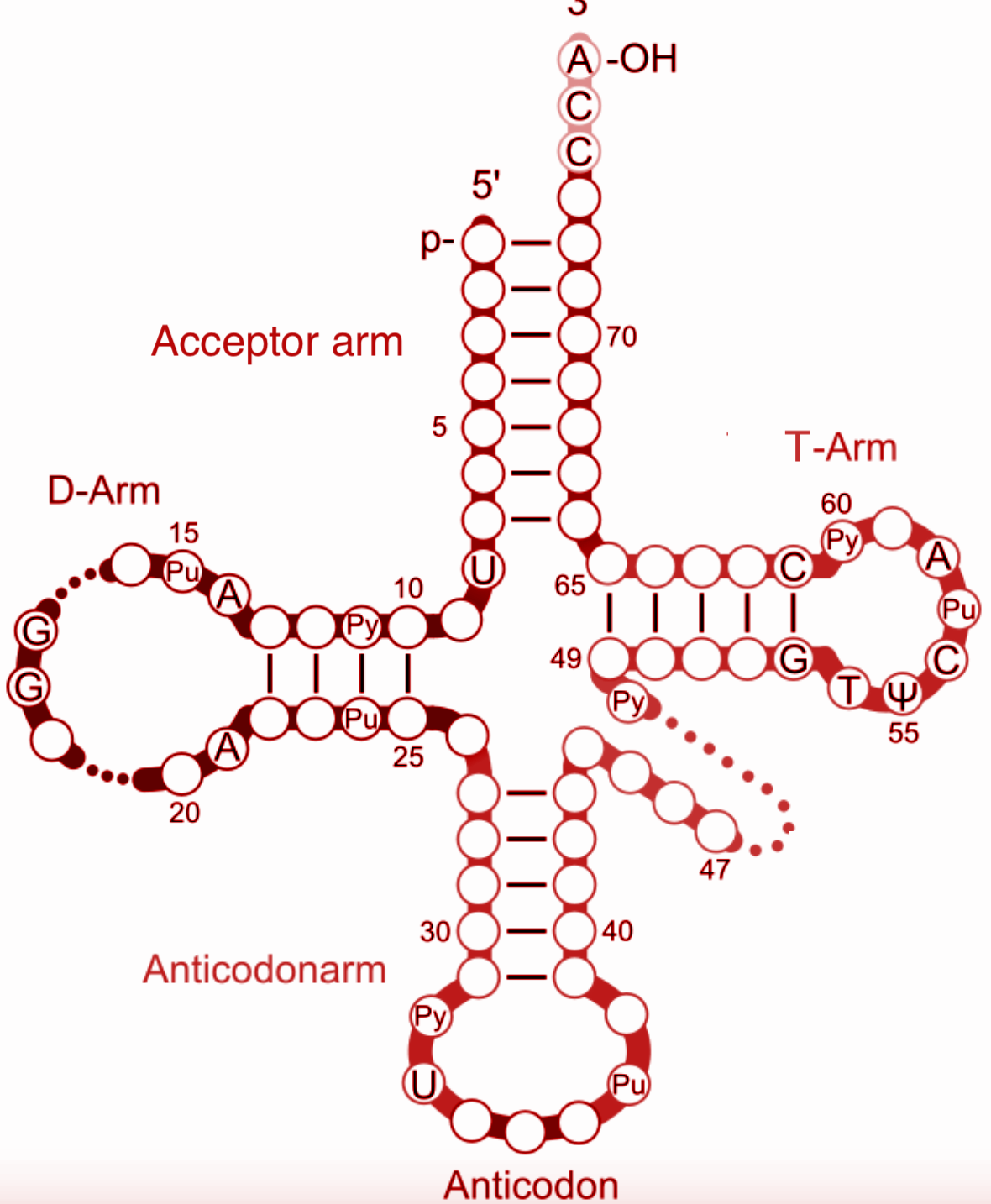
| Secondary Structure (Cloverleaf) | Folded with 4 main arms/loops: Acceptor arm, D arm, Anticodon arm, TΨC arm; variable loop present | Exam diagram; main for MCQs |
| Tertiary Structure | Further folded into L-shape by H-bonds | Active during translation |
| Charging (Aminoacylation) | Amino acid binds to 3' CCA tail (acceptor arm) | Enzyme: Aminoacyl tRNA synthetase |
Mechanism of Short Notes Of Biology For NEET tRNA Structure
The basic mechanism involves:
1. tRNA folds into a cloverleaf-shaped secondary structure.
2. The acceptor arm at 3' CCA tail binds the correct amino acid.
3. The anticodon arm has a complementary sequence matching mRNA codons.
4. During translation, tRNA “reads” the genetic code and brings the correct amino acid to the ribosome.
5. Variable loop and DHU, TΨC arms help proper recognition and stability.
Key Features and Functions
- The tRNA structure for NEET has four main arms: acceptor, D, anticodon, and TΨC; a variable loop may vary in length.
- Acts as an “adapter molecule” connecting genetic information (codons) to proper amino acids.
- Essential for translation (protein synthesis) in ribosomes.
- Each tRNA carries only one specific amino acid, determined by its anticodon sequence.
- No tRNA exists for stop codons.
Practice Questions
- What is the role of tRNA structure in the human body?
- Explain the steps involved in tRNA charging (aminoacylation).
- Draw and label a cloverleaf diagram of tRNA and mark all arms and loops.
- How does tRNA differ from mRNA and rRNA?
Common Mistakes to Avoid
- Confusing tRNA structure with mRNA or rRNA structure.
- Forgetting the CCA tail at the 3' end (amino acid attachment site) in diagrams.
- Overlooking the variable loop's presence and its position.
- Mixing up the position of the anticodon loop vs. the acceptor arm in labeled diagrams.
Real-World Applications
The concept of Short Notes Of Biology For NEET tRNA Structure is used in fields like medicine, molecular biology, and genetic engineering. Understanding tRNA helps explain how antibiotics may affect protein synthesis, how mutations in tRNA genes cause diseases, and how synthetic biology uses tRNA for making new proteins. Vedantu helps students relate such exam topics to practical medical and research examples.
In this article, we explored Short Notes Of Biology For NEET tRNA Structure, its structure, essential arms and loops, main function, real-world significance, and common diagram-based NEET questions. To learn more and build mastery, keep practicing with Vedantu and revise concepts using labeled diagrams and MCQs.
FAQs on tRNA Structure Short Notes for NEET Biology with Diagram
1. What is the structure of tRNA in NEET short notes?
The **tRNA structure** is typically 76-90 nucleotides long and folds into a **cloverleaf secondary structure** comprising an **acceptor arm**, **D arm (DHU loop)**, **anticodon loop**, **TΨC arm**, and a **variable loop**. Its **tertiary structure** forms an **L-shaped 3D conformation** stabilized by hydrogen bonds, crucial for its role in **protein synthesis**.
2. What is the short note on charging of tRNA?
**Charging of tRNA**, or **aminoacylation**, is the process where a specific **amino acid** is attached to the **3' end (CCA tail)** of the tRNA by the enzyme **aminoacyl tRNA synthetase**. This step is essential for **translation** as it prepares tRNA to deliver the correct amino acid corresponding to the mRNA codon.
3. What does the variable loop do in tRNA?
The **variable loop** in tRNA varies in size (3-21 nucleotides) and plays a key role in the **recognition of specific tRNA molecules** by enzymes and ribosomal components during translation. It helps distinguish between different types of tRNAs, adding specificity to the protein synthesis process.
4. How do I remember the cloverleaf diagram of tRNA for NEET?
To remember the **cloverleaf diagram** of tRNA for NEET:
1. Visualize the four arms: **Acceptor arm** (with CCA tail), **D arm**, **anticodon arm**, and **TΨC arm**.
2. Recall the **anticodon loop** pairs with mRNA codon.
3. Use mnemonic aids associating loop names with their functions.
4. Practice drawing the labelled diagram multiple times for quick recall.
5. What is the difference between tRNA, mRNA, and rRNA?
**tRNA** is a cloverleaf-shaped RNA that carries specific **amino acids** to the ribosome during **protein synthesis**. **mRNA** is a linear, single-stranded RNA that carries the **genetic code** from DNA to ribosomes. **rRNA** is a structural and functional component of ribosomes, facilitating the **assembly of amino acids into proteins**.
6. Why do students confuse the acceptor arm and anticodon arm in diagrams?
Students often confuse the **acceptor arm** and **anticodon arm** because both are key functional regions in tRNA but are located at opposite ends. The **acceptor arm** is at the **3’ end (CCA tail)** where the amino acid attaches, while the **anticodon arm** contains the **anticodon loop** that pairs with mRNA codons. Focusing on their **distinct functions and positions** helps reduce confusion.
7. How can I avoid missing marks on tRNA’s variable loop in MCQ diagrams?
To avoid missing marks on the **variable loop**:
1. Memorize that the variable loop lies between the **anticodon loop** and **TΨC loop**.
2. Note its variable size compared to other loops.
3. Practice labelled diagram questions emphasizing the variable loop location.
4. Understand its role to better recognize it in MCQs.
8. What’s a common error in remembering tRNA amino acid attachment site?
A common error is confusing the **3' terminal CCA tail** on the **acceptor arm** with other loops. The correct amino acid attachment site is the **3' hydroxyl group** of the **acceptor stem’s CCA sequence**. Remembering this terminal location and its unique sequence prevents mistakes.
9. Why do some confuse tertiary and secondary tRNA structure in NEET?
Confusion arises because the **secondary structure** of tRNA looks like a flat **cloverleaf**, while the **tertiary structure** folds into an **L-shape 3D conformation** stabilized by hydrogen bonds. NEET questions often show 2D diagrams, so clarifying that the tertiary form is the functional 3D shape in cells helps distinguish the two.
10. What is the role of aminoacyl tRNA synthetase in translation?
**Aminoacyl tRNA synthetase** is an enzyme that specifically attaches an **amino acid** to its corresponding **tRNA molecule** during the **charging** process. This ensures that the tRNA delivers the correct amino acid to the ribosome, maintaining the fidelity of **protein synthesis**.
11. How does the anticodon loop function in translation?
The **anticodon loop** contains a sequence of three nucleotides (**anticodon**) that pairs complementary to the **mRNA codon** during translation. This specific base pairing ensures the incorporation of the correct amino acid into the growing polypeptide chain.
12. Why are stop codons not recognised by any tRNAs?
**Stop codons** (UAA, UAG, UGA) do not have corresponding **tRNAs** because they signal the termination of **translation**. Instead, they are recognized by **release factors**, causing the ribosome to release the completed polypeptide chain.