How Do Lagging and Leading Strands Function During DNA Replication?
The concept of difference between lagging and leading strand in DNA replication is essential in biology and helps explain real-world biological processes and exam-level questions effectively, especially for students preparing for CBSE, NEET, and other competitive exams.
Understanding the Difference Between Lagging and Leading Strand
Difference between lagging and leading strand in DNA replication refers to the distinct ways the two new DNA strands are built during DNA synthesis. In the replication fork, the leading strand is synthesized continuously in the 5' to 3' direction, while the lagging strand is made discontinuously as small fragments called Okazaki fragments. This concept is important in understanding the structure of DNA, the central dogma of molecular biology, and the basic mechanism of DNA replication.
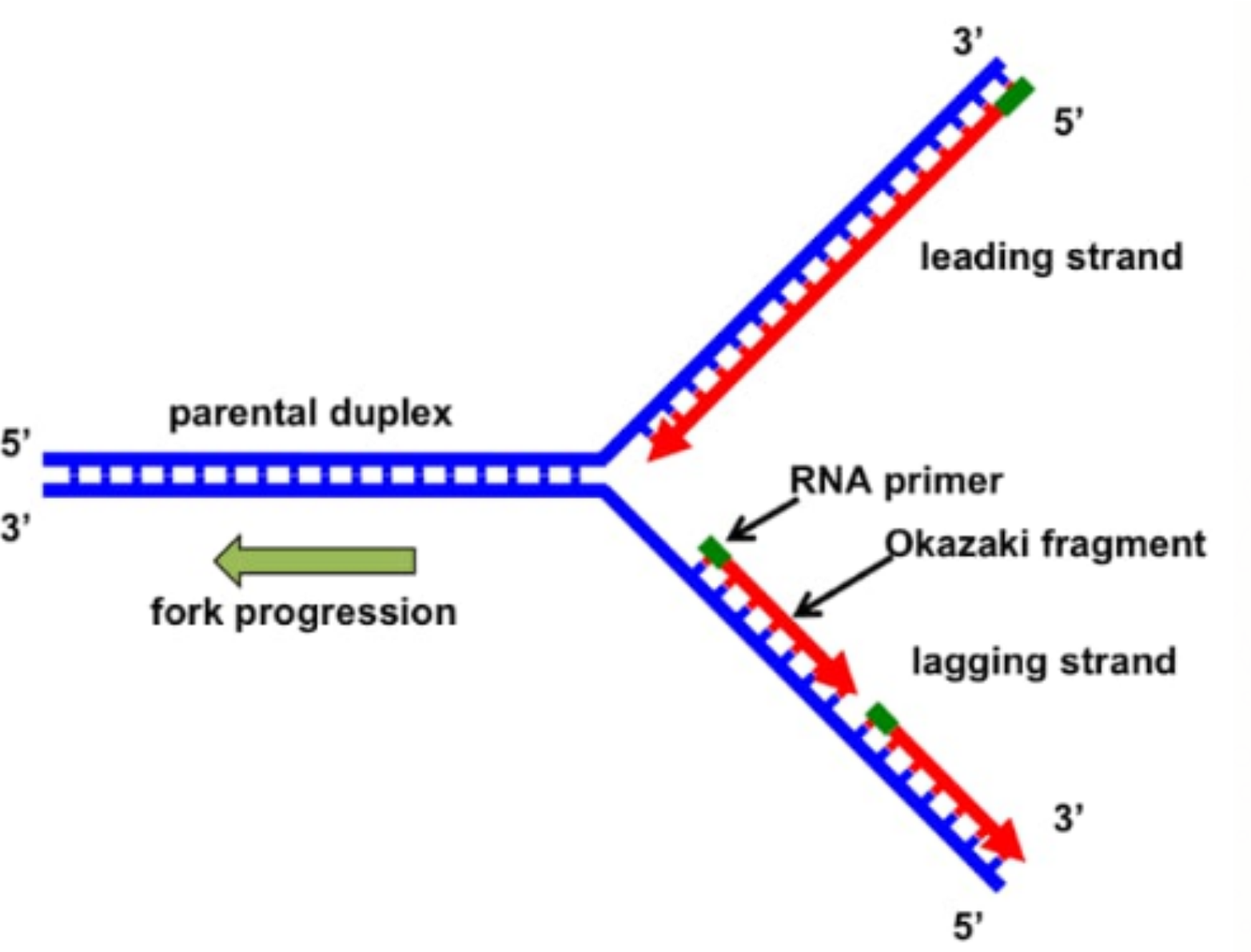
Mechanism of Difference Between Lagging and Leading Strand
The basic mechanism involves the action of DNA polymerase at the replication fork. Since DNA polymerase can only add nucleotides in the 5' to 3' direction, one strand is formed smoothly (leading), while the other is built in short bursts (lagging). The lagging strand needs repeated priming, and the resulting Okazaki fragments are later joined by DNA ligase.
Here’s a helpful table to understand the difference between lagging and leading strand better:
Difference Between Lagging and Leading Strand Table
| Feature | Leading Strand | Lagging Strand |
|---|---|---|
| Direction of Synthesis | 5' to 3' towards replication fork | 5' to 3' away from replication fork |
| Manner of Synthesis | Continuous | Discontinuous (forms Okazaki fragments) |
| Priming Requirement | Single RNA primer needed | Multiple RNA primers needed |
| Enzyme Actions | DNA polymerase moves smoothly | DNA polymerase detaches and reattaches |
| Associated Fragments | No Okazaki fragments | Okazaki fragments joined by DNA ligase |
Step-by-Step Process of DNA Replication: Leading vs Lagging Strand
Let’s understand the synthesis steps for each strand in DNA replication:
- Replication fork opens: DNA helicase unwinds the double helix.
- Leading strand: DNA polymerase starts adding nucleotides in the 5' to 3' direction continuously, following the fork.
- Lagging strand: Primase adds multiple RNA primers along the strand, and DNA polymerase synthesizes short Okazaki fragments between primers.
- Okazaki fragments: These fragments are synthesized as the fork exposes new template regions.
- Fragment joining: DNA ligase connects Okazaki fragments to form a continuous lagging strand.
Similarities and Common Misconceptions
- Both strands are synthesized in the 5' to 3' direction by DNA polymerase.
- Each uses common enzymes such as DNA polymerase, primase, and ligase.
- Common Mistake: Believing the lagging strand is made 3' to 5'. In reality, both are built 5' to 3'.
- Some students mix up which strand is continuous and which has Okazaki fragments.
Practice Questions
- What is the primary difference between lagging and leading strands in DNA replication?
- Explain how Okazaki fragments are formed and joined.
- List three differences between the lagging and leading strand.
- Label the leading and lagging strands on a diagram of the replication fork.
- Which strand is synthesized in a discontinuous manner and why?
Common Mistakes to Avoid
- Confusing difference between lagging and leading strand with template and coding strands.
- Assuming DNA polymerase can synthesize both strands continuously.
- Not mentioning the role of primers and DNA ligase in lagging strand synthesis.
Real-World Applications
The concept of difference between lagging and leading strand is widely used in genetics, biotechnology, and medical diagnostics. Understanding DNA replication helps in the study of genetic diseases, designing DNA tests, and research in fields like gene cloning. Vedantu helps students connect such topics to real-world biological research and modern healthcare advances.
In this article, we explored difference between lagging and leading strand, its key processes, real-life significance, and how to solve questions based on it. To learn more and build confidence, keep practicing with Vedantu.
Quick Revision Table
| Leading Strand | Lagging Strand |
|---|---|
| Synthesized continuously, single primer, no Okazaki fragments | Synthesized discontinuously, multiple primers, forms Okazaki fragments |
Explore Related Topics
- DNA Structure
- Nucleic Acid
- Central Dogma of Molecular Biology
- DNA Replication
- Difference Between Prokaryotic and Eukaryotic DNA Replication
- Okazaki Fragments
- Gene Definition
- RNA and Transcription
- Nucleotide
- Difference Between Gene and Chromosome


FAQs on Lagging vs Leading Strand: Differences in DNA Replication
1. What is the main difference between the leading and lagging strand in DNA replication?
The primary difference lies in their mode of synthesis. The leading strand is synthesized continuously in a single piece, moving towards the replication fork. In contrast, the lagging strand is synthesized discontinuously in short segments called Okazaki fragments, moving away from the replication fork. This occurs because DNA polymerase can only add nucleotides in the 5' to 3' direction.
2. Why are Okazaki fragments formed only on the lagging strand?
Okazaki fragments are necessary because the two parental DNA strands are antiparallel. One template strand runs 3' to 5', allowing the new (leading) strand to be synthesized continuously in the 5' to 3' direction. However, the other template (lagging) strand runs 5' to 3'. To maintain the mandatory 5' to 3' synthesis direction, DNA polymerase must create the new strand in short, discontinuous bursts as the DNA unwinds, resulting in Okazaki fragments.
3. If both strands are synthesized 5' to 3', how is their direction relative to the replication fork different?
This is a common point of confusion. While the biochemical process of adding nucleotides is always 5' to 3' for both strands, their overall growth direction differs. The leading strand is synthesized continuously towards the advancing replication fork. The lagging strand is also synthesized in a 5' to 3' direction, but each Okazaki fragment is made in the direction away from the replication fork. The polymerase essentially works backwards on the lagging strand template.
4. What are the key enzymes involved in the synthesis of the lagging strand compared to the leading strand?
Both strands use a core set of enzymes like Helicase (to unwind DNA), Primase (to create RNA primers), and DNA Polymerase III (to synthesize DNA). However, the lagging strand has additional requirements:
- It needs multiple RNA primers, one for each Okazaki fragment, whereas the leading strand typically needs only one.
- It requires DNA Polymerase I to remove the RNA primers and replace them with DNA nucleotides.
- It uniquely requires the enzyme DNA Ligase to join the Okazaki fragments into a continuous strand by forming phosphodiester bonds.
5. How does the structure of the replication fork dictate the existence of a leading and a lagging strand?
The Y-shaped replication fork is the site of active DNA unwinding. This structure simultaneously exposes two template strands with opposite polarities (one 3'→5' and the other 5'→3'). Since DNA Polymerase can only read a template in the 3'→5' direction to synthesize a new 5'→3' strand, it can move along one template continuously. This creates the leading strand. For the other template, the enzyme must wait for a segment to unwind and then synthesize a fragment backwards, leading to the discontinuous formation of the lagging strand.
6. What is the ultimate fate of Okazaki fragments after they are synthesized?
Okazaki fragments are temporary structures. Once synthesized, the RNA primers at the beginning of each fragment are removed by the exonuclease activity of DNA Polymerase I. The same enzyme then fills these gaps with the correct DNA nucleotides. Finally, the enzyme DNA Ligase creates a phosphodiester bond to seal the nick between the fragments, creating a single, continuous, and stable DNA strand.
7. Could the leading strand ever be synthesized discontinuously?
No, by definition, the leading strand is the one that is synthesized continuously. Its continuous nature is a direct consequence of its template strand having the correct 3' to 5' orientation relative to the movement of the replication fork. If synthesis were to become discontinuous, it would be functionally defined as a lagging strand.
8. What is the real-world importance of understanding the difference between leading and lagging strand synthesis?
Understanding this fundamental mechanism is crucial in various fields of biology and medicine. For example:
- Biotechnology: Techniques like the Polymerase Chain Reaction (PCR) for DNA amplification are based on the principles of directional synthesis by DNA polymerase.
- Genetics: It helps explain how errors during replication can lead to mutations and genetic diseases.
- Drug Development: Some antiviral and anticancer drugs work by targeting and inhibiting specific enzymes in the replication process, like DNA polymerase or ligase.










